Breadcrumb
- Home
- Current Projects
Current Projects
RNA Polymerase II Elongation Control
The Price Lab is interested in all processes that take place during the elongation phase of transcription by RNA polymerase II. This includes the elongation control process which is mediated by P-TEFb, termination by TTF2, and the RNA processing events that are functionally coupled to transcription elongation. We are also involved in understanding how viruses utilize the elongation control system during active infections and latency and how Pol II influences and is influenced by chromatin.

The big picture
The human genetic material is crammed into the tiny nucleus in every cell and by default held in a highly compacted and repressed state. Transcription of genes by Pol II is accomplished by initiation (green arrows) at promoters and in enhancers in both sense and divergent directions. Every initiation event leads to promoter proximal pausing and this allows the promoter region to be held in an active state (bright yellow). Five factors have been identified that contribute to promoter proximal pausing: DSIF, NELF, TFIIS, Gdown1, and the Gdown1 negative accessory factor, GNAF. Paused Pol II either terminates (red arrows) or transitions into productive elongation which allows transcription of the gene at 4,000 nt/min. P-TEFb controls the fate of the paused Pol II by phosphorylating DSIF which triggers the loss of NELF and allows acquisition of downstream productive elongation factors such as the PAF1 complex. The goal of our work is to understand the mechanisms involved in Pol II elongation control. Some of our current projects are described below.
Control of P-TEFb
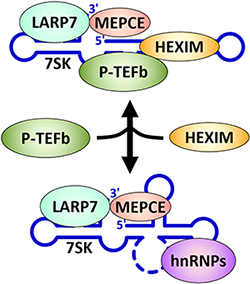
P-TEFb is regulated by reversible interaction with the 7SK snRNP. Association with HEXIM1 or HEXIM2 in the 7SK snRNP inhibits P-TEFb and maintains a large store of P-TEFb that can be released when and potentially where it is needed. LARP7 and MePCE are constitutive components of the 7SK snRNP that maintain stability of the complex and are likely involved in the regulated release of P-TEFb. Studies are underway to uncover the mechanisms that control release and re-sequestration of P-TEFb into the 7SK snRNP.
Viral models of transcription
The Price lab exploits viral models to understand Pol II elongation control. We showed over the last 20 years how an HIV protein Tat takes over the control of P-TEFb, bypassing the normal cellular control process, by directly recruiting P-TEFb to the integrated viral promoter. We are currently working with Jeff Meier (Internal Medicine) to examine the transcriptional life cycle of the human cytomegalovirus (HCMV). HCMV is a human-restricted betaherpesvirus that infects over half of all Americans and causes a variety of serious diseases. It surpasses Zika as the most common infectious cause of birth defects. Its genome consists of about a quarter of a million bps and it uses, in part, the host Pol II transcription machinery to express hundreds of viral genes. We are examining the importance of Pol II elongation control in the expression of the viral genome and how the virus affects transcription of host genes. This knowledge is necessary to derive future therapies that target viral transcription.
Promoter-proximal pausing
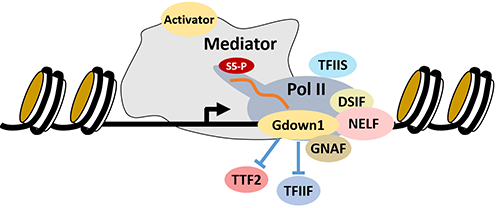
The sequence of events following initiation by Pol II is a highly orchestrated exchange of factors that are in part directed by post-translational modifications of Pol II and the factors. One of the steps is promoter-proximal pausing which is a required intermediate to productive elongation. Most of the pausing factors have been identified, but one of the important features of promoter-proximal pausing, the half-life of the complex, requires further study. Although some studies suggest that the paused complex is stable for minutes or hours, others suggest that termination occurs quickly (in less than a minute). The timing of the flux of Pol II through the cycle of initiation, pausing and termination is critical to understand how pausing affects the function of promoters.
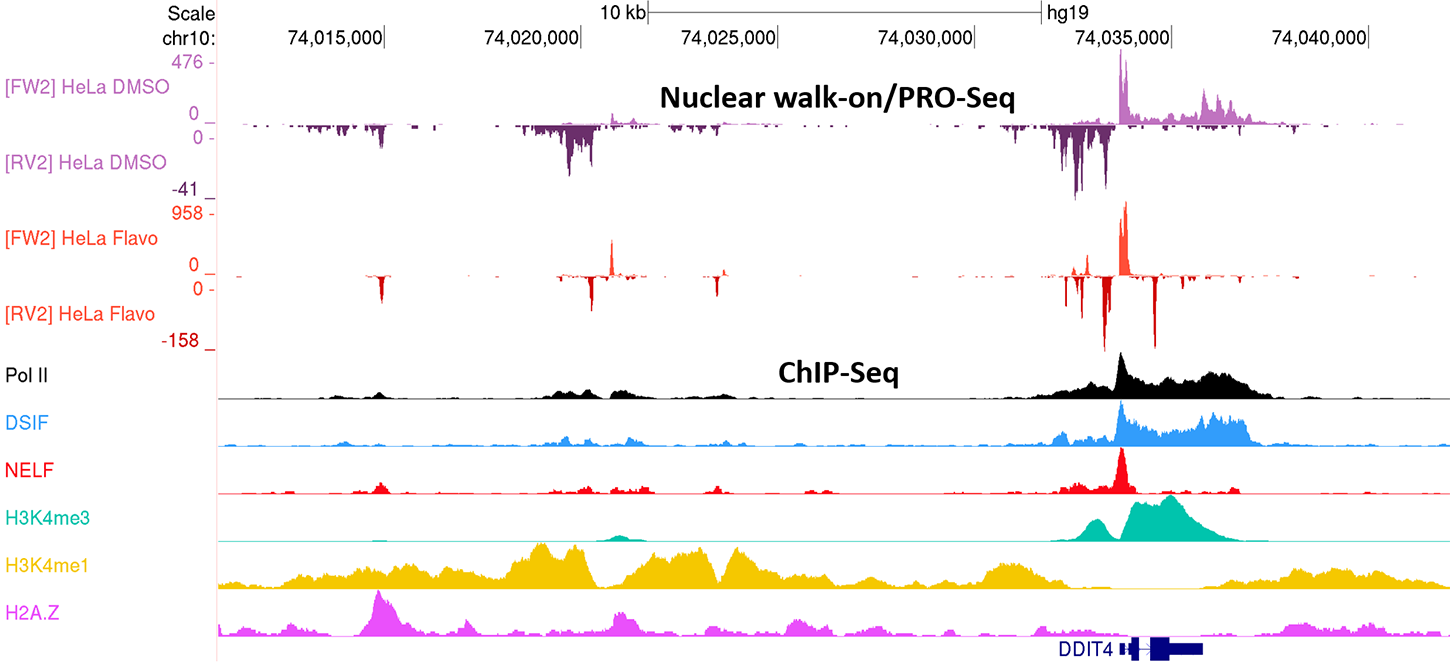
Transcription through chromatin
The Pol II transcription machinery must interact appropriately with the nucleosomes which compact human genomic DNA to achieve the patterns of gene expression that drive development and regulated responses to changing cellular environments. Chromatin structure is dynamic, and both responds to and affects transcription. In collaboration the Don Luse (Cleveland Clinic), we are examining the interplay of chromatin and Pol II transcription. We have developed a method to globally analyze Pol II transcription that relies upon the use of nuclei that are rapidly isolated from cells. A nuclear “walk-on” assay provides a quantitative measure of the amount of paused Pol II in cells as well as the length of engaged transcripts. Coupling the walk-on assay with PRO-Seq we can obtain nucleotide resolution of both the 5′ and 3′ ends of nascent transcripts. This assay together with ChIP-Seq is being used to analyze cells that have been manipulated with small molecule inhibitors, specific protein degraders, siRNA knockdown of specific proteins and RNAs, and CRISPR modified genomes to tease out the how promoter proximal pausing and nucleosomes impact each other. In addition, we have begun to utilize the isolated nuclei as native chromatin templates for de novo initiation in vitro. These approaches should be able to resolve many of the remaining questions concerning transcription through nucleosome including, how nucleosomes and Pol II pausing and productive elongation influence each other.